Bacground: because the transcriptome data I recently worked with is highly related with immune. So I want to dig out something about immune.
When I viewed RNAseq analysis notes from Tommy Tang, I found ImReP. It’s a tool designed for profiling TCR/BCR repertoire in regular RNA-seq data. That was something I nerver heard before.
Then I found more tools through the paper of ImRep [1]. They are:
And the paper of ImRep introduce them as follows:
TRUST and TraCeR do not support the analysis of BCR sequences and were excluded from the comparison based for the IGH data. iSSAKE is no longer supported and was not recommended for use. Unfortunately, we obtained empty output after running V’DJer, and increasing coverage in the simulated data did not solve the problem. Alternative approaches, such as IMSEQ, cannot be applied directly to RNA-Seq reads because they were originally designed for targeted sequencing of B or T cell receptor loci. Thus, to independently assess and compare accuracy with ImReP, we only ran IMSEQ with the simulated reads derived from BCR or TCR transcripts (Figure S1). Scripts and commands to run all tools used in this study are provided in the Extended Experimental Procedures and are available online at https://github.com/smangul1/Profiling-adaptive-immune-repertoires-across-multiple-humantissues-by-RNA-Sequencing. ImReP consistently outperformed existing methods on IGH data in both recall and precision rates for the majority of simulated parameters. ImReP and MiXCR show similar performance on TCRA data and outperform other methods. Notably, ImReP was the only method with acceptable performance on IGH data at 50bp read length, reconstructing with a higher precision rate significantly more CDR3 clonotypes than other methods.
Because I only have regular RNA-seq data (non-enriched and/or randomly-shred ©DNA libraries), ImRep and MiXCR were the only two software I want to try.
MiXCR
Intro
MiXCR is a universal framework that processes big immunome data from raw sequences to quantitated clonotypes. MiXCR efficiently handles paired- and single-end reads, considers sequence quality, corrects PCR errors and identifies germline hypermutations. The software supports both partial- and full-length profiling and employs all available RNA or DNA information, including sequences upstream of V and downstream of J gene segments. (https://mixcr.readthedocs.io/en/latest/)
MiXCR has very nice docs: https://mixcr.readthedocs.io/en/latest. See them for full instructions.
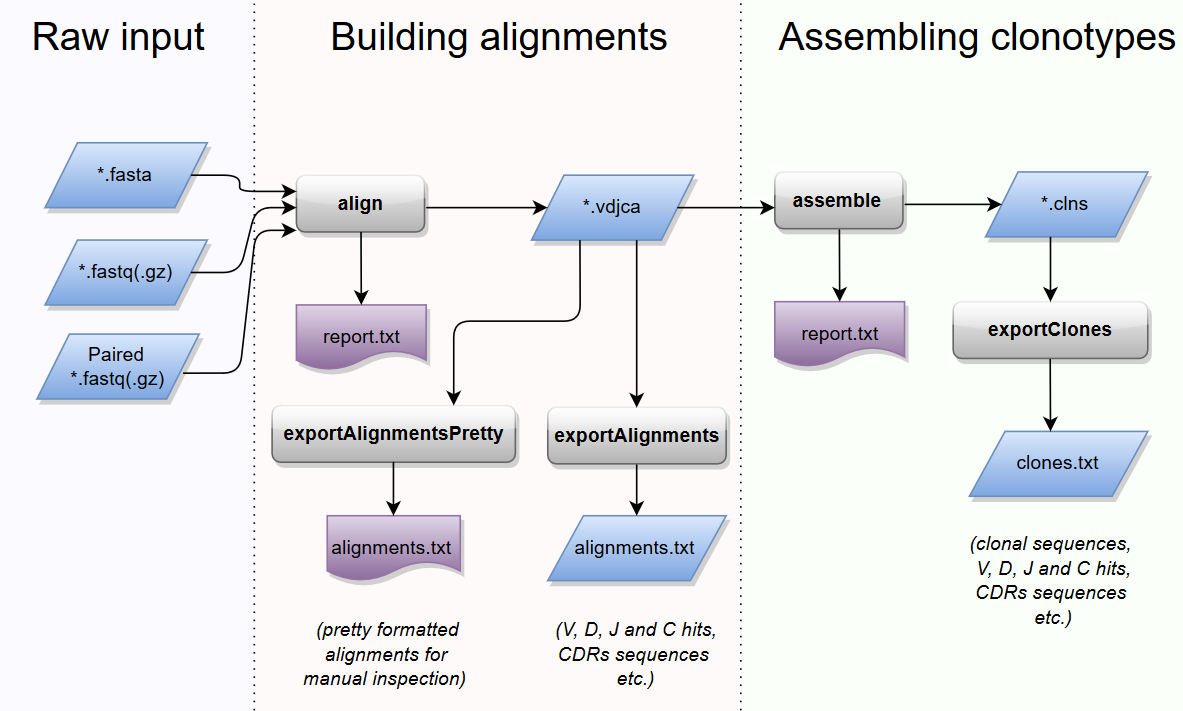
Typical MiXCR workflow consists of three main processing steps:
- align: align sequencing reads to reference V, D, J and C genes of T- or B- cell receptors
- assemble: assemble clonotypes using alignments obtained on previous step (in order to extract specific gene regions e.g. CDR3)
- export: export alignment (
exportAlignments) or clones (exportClones) to human-readable text file
Enriched RepSeq Data
Here is a very simple usage example that will extract repertoire data (in the form of clonotypes list) from raw sequencing data of enriched RepSeq library:
2
3
mixcr assemble -r log.txt alignments.vdjca clones.clns
mixcr exportClones clones.clns clones.txt
this will produce a tab-delimited list of clones (
clones.txt) assembled by their CDR3 sequences with extensive information on their abundances, V, D and J genes, mutations in germline regions, topology of VDJ junction etc.
Repertoire extraction from RNA-Seq
MiXCR is equally effective in extraction of repertoire information from non-enriched data, like RNA-Seq or WGS. This example illustrates usage for RNA-Seq:
2
3
4
mixcr assemblePartial alignments.vdjca alignment_contigs.vdjca
mixcr assemble -r log.txt alignment_contigs.vdjca clones.clns
mixcr exportClones clones.clns clones.txt
Install
- download the latest stable
MiXCRbuild from release page
1 | unzip mixcr-2.1.9.zip && cd mixcr-2.1.9 |
Run
Pipelines from https://github.com/milaboratory/mixcr and http://mixcr.readthedocs.io/en/latest/rnaseq.html are not exactly the same. I used the latter.
1 | path2MiXCR=$TOOLDIR/mixcr-2.1.9/mixcr |
And the output looks like this:
1 | head -n 3 clones.txt |
No ideas what to do next… I will try some post-analysis tools to explore the clonetypes.
ImRep
Intro
ImReP is a method for rapid and accurate profiling of the adaptive immune repertoires from regular RNA-Seq data.
Install
1 | git clone https://github.com/mandricigor/imrep.git && cd imrep |
Run
Then I was caught in an embarrassing situation.
ImRep now is designed to handle two cases:
- When you have saved mapped and unmapped reads in one
BAMfile,ImRepcan accept oneBAMas input.
Given the bam file with mapped and unmapped reads, you can run ImReP using this command.
python imrep.py --bam example/toyExample.bam example/toyExample.cdr3
- When you forgot to save unmapped reads,
ImRepcan acceptBAMfile with mapped reads and all rawFASTQfiles as input.
Forgot to save unmapped reads, we got you covered. Use
--digGoldand-aoptions. For example:
python imrep.py --digGold -a example/toyExample_allReads.fastq example/toyExample_onlyMapped.bam example/toyExample.cdr3
I’ve aligned the FASTQ files to genome with STAR, and saved unmapped reads in FASTQ format (using --outReadsUnmapped Fastx).
And the author said:
Some mapping tools produce partially-mapped reads (i.e. STAR). In case read is mapped to BCR or TCR genes and is partially mapped to V or J gene, such read may be used to assemble full-length CDR3 sequences. Considering only unmapped reads will result in missing such reads.
So the first case doesn’t suit me, I have to follow the second.
And the questions are:
- Can I feed
ImRepwith theBAMand unmapped reads? not all raw reads. - It seems
ImReponly accepts one singleFATSQas input, should I cat twoFASTQof pair-end data? Or it just works for single-end data?
I reported a issue to the author: unmapped reads in fastq/fasta format and pair-end data.
And he suggested:
Please merge PE into one file. Also to use --digGold, you need to provide original reads, not the unmapped reads. Please let me know how it goes. If this doesn’t’ work for you, we can implement the option to allow to supply bam with mapped and FASTQ with unmapped (this is on our TODO list anyway). Thanks, Serghei
I should run STAR with --outSamUnmapped Within option hereafter. And I’ll not plan to re-align the reads for now.
Moreover, using MiXCR, I can use post-analysis tools such as VDJtools easily.
Change notes
- 20180324: create the note.
Mangul S, Mandric I, Yang HT, Strauli N, Montoya D, Rotman J, Wey WVD, Ronas JR, Statz B, Zelikovsky A, et al. Profiling adaptive immune repertoires across multiple human tissues by RNA Sequencing. bioRxiv. 2017 Mar 25:089235. doi:10.1101/089235 ↩︎
Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: software for comprehensive adaptive immunity profiling. Nature Methods. 2015;12(5):380–381. doi:10.1038/nmeth.3364 ↩︎
Li B, Li T, Wang B, Dou R, Pignon J-C, Choueiri TK, Signoretti S, Liu JS, Liu XS. Ultrasensitive detection of TCR hypervariable region in solid-tissue RNA-seq data. bioRxiv. 2016 Sep 5:073395. doi:10.1101/073395 ↩︎
Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nature Methods. 2016;13(4):329–332. doi:10.1038/nmeth.3800 ↩︎
Mose LE, Selitsky SR, Bixby LM, Marron DL, Iglesia MD, Serody JS, Perou CM, Vincent BG, Parker JS. Assembly-based inference of B-cell receptor repertoires from short read RNA sequencing data with V’DJer. Bioinformatics. 2016;32(24):3729–3734. doi:10.1093/bioinformatics/btw526 ↩︎
Strauli NB, Hernandez RD. Statistical inference of a convergent antibody repertoire response to influenza vaccine. Genome Medicine. 2016;8:60. doi:10.1186/s13073-016-0314-z ↩︎
Warren RL, Nelson BH, Holt RA. Profiling model T-cell metagenomes with short reads. Bioinformatics (Oxford, England). 2009;25(4):458–464. doi:10.1093/bioinformatics/btp010 ↩︎