Reliable cancer gene collections are useful in cancer research. Here I list several such resources.
These sets can be devided into two categories: mutation- or data-based, and literature- or knowledge-based.
Mutation-based
COSMIC Cancer Gene Census (CGC)
- Paper: The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers
- Web page: https://cancer.sanger.ac.uk/census
The Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census (CGC) is an expert-curated description of the genes driving human cancer. The figure following shows the curation process.
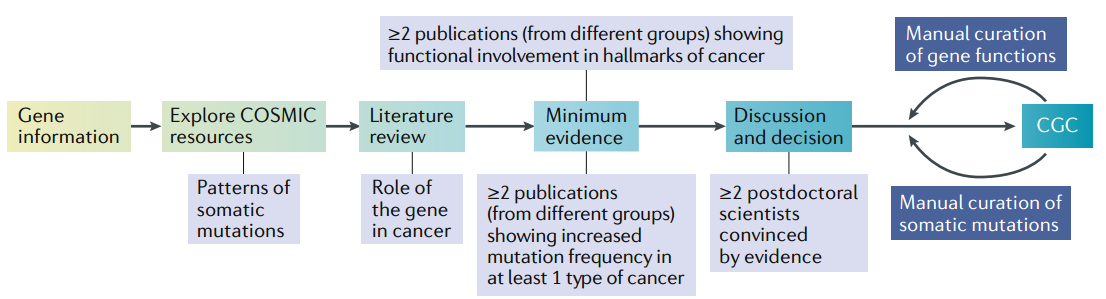
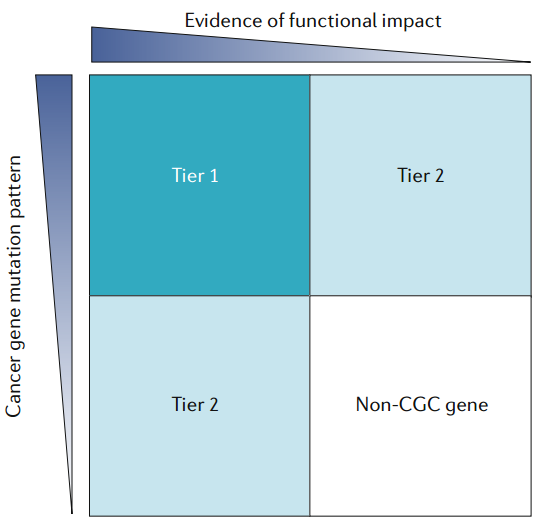
It comprises two tiers.
- To classify as Tier 1, a gene must possess a documented and reproducible activity relevant to cancer, along with evidence of mutations in cancer that change the activity of the gene product in a way that promotes oncogenic transformation.
- Included in Tier 2 are genes with mutation patterns typical for oncogenes or TSGs but that have less well-established functional evidence in the scientific literature. Similarly, genes with strong published evidence for a function in cancer but unclear mutation patterns or genes known to be dysregulated solely by epigenetic means (for example, by changes to promoter methylation) are also included in Tier 2.
Comments
The Cancer Gene Census (CGC) uses data from the Catalogue of Somatic Mutations in Cancer (COSMIC) to list known oncogenes and tumor suppressors.
20/20 rule
- Paper: Cancer Genome Landscapes
The authors used mutation patterns to classify genes of the COSMIC database into oncogenes and tumor supressor genes.
- To be classified as an oncogene, we simply require that >20% of the recorded mutations in the gene are at recurrent positions and are missense.
- To be classified as a tumor suppressor gene, we analogously require that >20% of the recorded mutations in the gene are inactivating.
Network of Cancer Genes (NCG)
- Paper: The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens
- Web page: http://ncg.kcl.ac.uk/
Genes in NCG were collected from 275 publications, including two sources of known cancer genes and 273 cancer sequencing screens of more than 100 cancer types from 34,905 cancer donors and multiple primary sites.
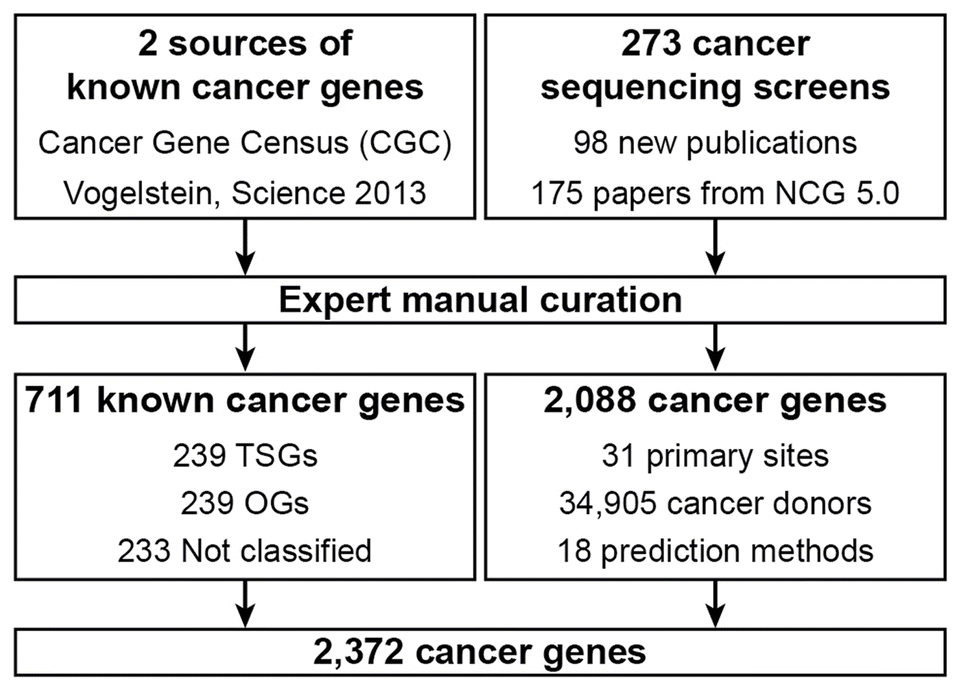
Comments
The Network of Cancer Genes builds upon the CGC and integrates a wide variety of additional contextual data, such as the frequency of mutations.
IntOGen
- Paper: IntOGen-mutations identifies cancer drivers across tumor types
- Web page: https://www.intogen.org/search
IntOGen-mutations is a Web platform used to identify cancer drivers across tumor types and to present the results of the systematic analysis of most currently available large data sets of tumor somatic mutations.
Comments
IntOGen uses data from large-scale sequencing projects (for example, the Cancer Genome Atlas (TCGA)) to collate the importance of cancer genes.
Candidate Cancer Gene Database (CCGD)
- Paper: The Candidate Cancer Gene Database: a database of cancer driver genes from forward genetic screens in mice
- Web page: http://ccgd-starrlab.oit.umn.edu/about.php
The Candidate Cancer Gene Database (CCGD) was developed to disseminate the results of transposon-based forward genetic screens in mice that identify candidate cancer genes. The purpose of the database is to allow cancer researchers to quickly determine whether or not a gene, or list of genes, has been identified as a potential cancer driver in a forward genetic screen in mice.
Literature-based
All manually curated databases face the overwhelming curation burden of expert curator time and costs necessary to stay up-to-date.
CancerMine
- Paper: CancerMine: a literature-mined resource for drivers, oncogenes and tumor suppressors in cancer
- Web page: http://bionlp.bcgsc.ca/cancermine/
- Github: https://github.com/jakelever/cancermine
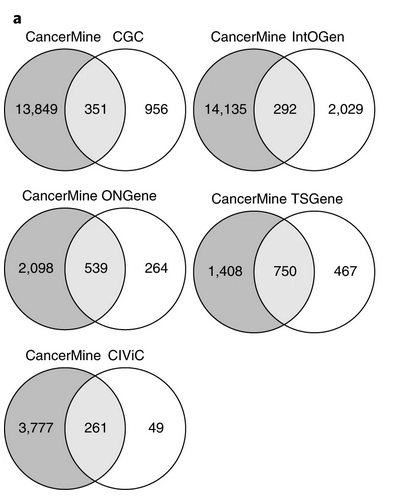
CancerMine a literature-mined database of drivers, oncogenes and tumor suppressors in cancer. The authors first manually annotated the drivers, oncogenes and tumor suppressors discussed in 1,500 sentences as training data. Then they trained a logistic regression classifier on word frequencies and semantic features. To lower the number of false positives, a high threshold was uded, resulting relatively high precision and low recall (average precision of 85.6% and recall of 29.4% across the three gene role types).
CancerMine contains substantially more cancer gene associations than other resources but has poor overlap with CGC and IntOGen. The authors explained this as “gene associations in the CGC and IntOGen are not mentioned in the literature”.
ONGene
- Paper: ONGene: A literature-based database for human oncogenes
- Web page: http://ongene.bioinfo-minzhao.org
ONGene is a database for oncogenes. The authors manually curated abstracts from PubMed (Dec 25th, 2015) and collected 803 human oncogenes (698 protein-coding genes and 105 non-coding genes.)
This database is simple and has high utility.
Comments
ONGene and TSGene list oncogenes and tumor suppressors but do not associate them with specific cancer types.
TSGene
- Paper: TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes
- Web page: http://bioinfo.mc.vanderbilt.edu/TSGene/
TSGene is a database for tumor suppressor genes. The authors manually curated abstracts from PubMed (25 April 2015) and collected 1217 human TSGs (1018 protein-coding genes and 199 non-coding genes.)
Comments
ONGene and TSGene list oncogenes and tumor suppressors but do not associate them with specific cancer types.
Clinical Interpretation of Variants in Cancer (CIViC)
- Paper: CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer
- Web page: https://civicdb.org/home
CIViC is an expert-crowdsourced knowledgebase for Clinical Interpretation of Variants in Cancer describing the therapeutic, prognostic, diagnostic and predisposing relevance of inherited and somatic variants of all types.
Related resources
Change log
- 20190602: create the note.